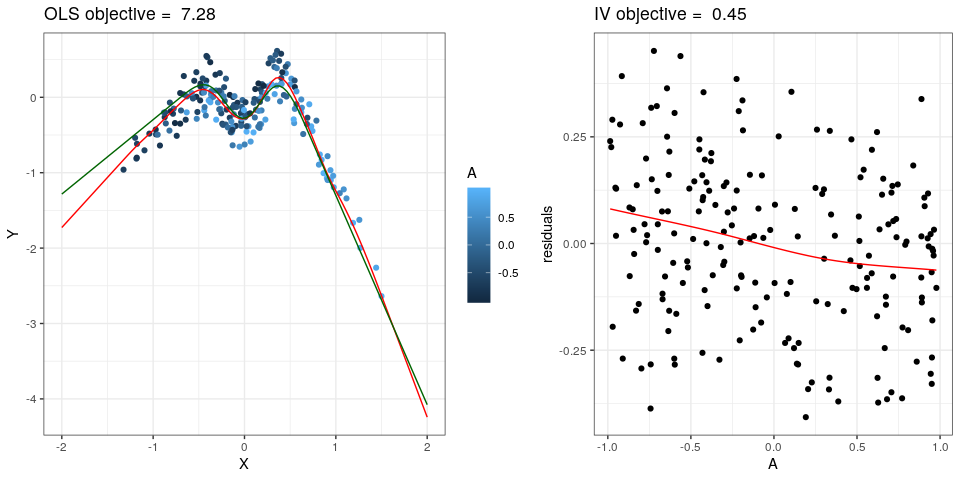
The NILE estimator can be used to estimate a nonlinear causal influence of a single predictor X on a real-valued response Y. It exploits an instrumental variable (IV) setting with a single exogenous variable A which serves as instrument. Predictions obtained from the NILE estimator are causal predictions in the sense that they predict the values of Y under interventions which break the dependence between X and possible confounders of (X, Y). For further details refer to the paper Christiansen et al. (2020 https://arxiv.org/abs/2006.07433).
Installation
You can install the the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("runesen/NILE")Example
This is a basic example which shows the idea behind NILE. Let us start by importing the NILE library.
Suppose that the true (unknown) functional relationship between X and Y is defined by a spline that linearly extrapolates outside the training data.
# true (unknown) functional relationship X -> Y
# (linearly extrapolating beyond "extrap")
fX <- function(x, extrap, beta){
bx <- splines::ns(x, knots = seq(from=extrap[1], to=extrap[2],
length.out=(n.splines.true+1))[
-c(1,n.splines.true+1)],
Boundary.knots = extrap)
bx%*%beta
}Let us define the data generating model.
# data generating model
n <- 200
n.splines.true <- 4
set.seed(2)
beta0 <- runif(n.splines.true, -1,1)
alphaA <- alphaEps <- alphaH <- 1/sqrt(3)
A <- runif(n,-1,1)
H <- runif(n,-1,1)
X <- alphaA*A + alphaH*H + alphaEps*runif(n,-1,1)
Y <- fX(x=X,extrap=c(-.7,.7), beta=beta0) + .3*H + .2*runif(n,-1,1)
x.new <- seq(-2,2,length.out=100)
f.new <- fX(x=x.new,extrap=c(-.7,.7), beta=beta0)
plot(X,Y, pch=20)
lines(x.new,f.new,col="#0072B2",lwd=3)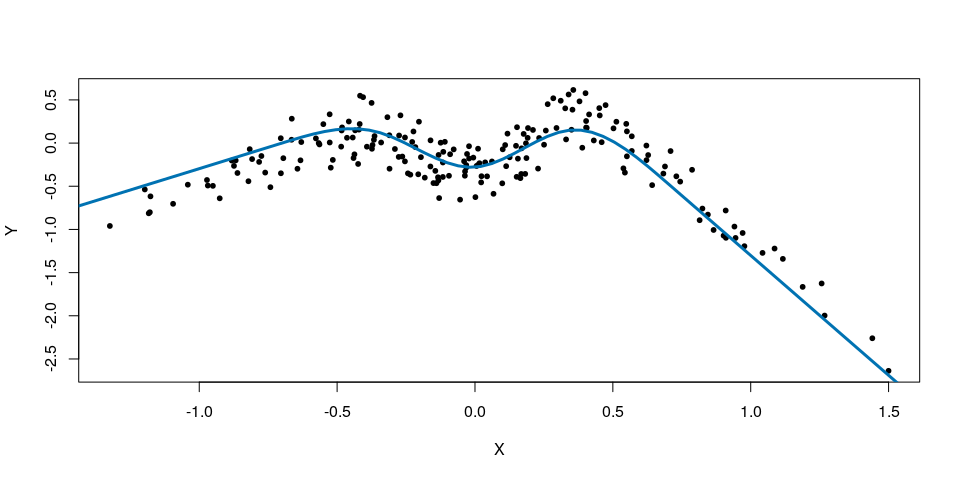
Given the observed dataset, let us now fit NILE.
fit <- NILE(Y, # response
X, # predictors (so far, only 1-dim supported)
A, # anchors (1 or 2-dim, although 2-dim is experimental so far)
lambda.star = "test", # (0 = OLS, Inf = IV, (0,Inf) =
# nonlinear anchor regression, "test" = NILE)
test = "tsls.over.ols",
intercept = TRUE,
df = 50, # number of splines used for X -> Y
p.min = 0.05, # level at which test for lambda is performed
x.new = x.new, # values at which predictions are required
plot=TRUE, # diagnostics plots
f.true = function(x) fX(x,c(-.7,.7), beta0), # if supplied, the
# true causal function is added to the plot
par.x = list(lambda=NULL, # positive smoothness penalty for X -> Y,
# if NULL, it is chosen by CV to minimize out-of-sample
# AR objective
breaks=NULL, # if breaks are supplied, exactly these
# will be used for splines basis
num.breaks=20, # will result in num.breaks+2 splines,
# ignored if breaks is supplied.
n.order=4 # order of splines
),
par.a = list(lambda=NULL, # positive smoothness penalty for fit of
# residuals onto A. If NULL, we first compute the OLS
# fit of Y onto X,
# and then choose lambdaA by CV to
# minimize the out-of-sample MSE for predicting
# the OLS residuals
breaks=NULL, # same as above
num.breaks=4, # same as above
n.order=4 # same as above
))
#> [1] "lambda.cv.a = 1.42932324587573"
#> [1] "lambda.cv.x = 0.0015933292003293"
#> [1] "lambda.star.p.uncorr = 1.45538091659546"